|
| ||||||||||
Applied and Environmental Microbiology, June 1999, p.
2606-2613, Vol. 65, No. 6
0099-2240/99/$04.00+0
Copyright © 1999, American Society for Microbiology. All rights
reserved.
School of Biological Sciences, University of Nebraska![]() Lincoln, Lincoln, Nebraska
68588-0666,1 and Department of Microbiology, Arizona College of
Osteopathic Medicine, Midwestern University, Glendale, Arizona 853082
Lincoln, Lincoln, Nebraska
68588-0666,1 and Department of Microbiology, Arizona College of
Osteopathic Medicine, Midwestern University, Glendale, Arizona 853082
Received 1 October 1998/Accepted 24 March 1999
| |
ABSTRACT |
|---|
UNL-1, a lytic virus of Pseudomonas aeruginosa, was observed to express a novel inducible DNA damage reactivation activity in UV-A-irradiated P. aeruginosa host cells. The expression of bacteriophage reactivation was quantified in hosts exposed to either UV-C or UV-A radiation. While reactivation of UV-C-damaged UNL-1 was not inducible in UV-C-irradiated host cells, an approximately 13-fold induction was observed in UV-A-irradiated host cells. When host cells were exposed to sunlight, reactivation of damaged UNL-1 virus increased eightfold. The UV-A induction of UNL-1 DNA damage reactivation was supported in hosts lacking recA gene function. This report is the first description of a recA-independent, UV-inducible virus DNA damage repair system. Our findings suggest that a combination of both host and virus DNA repair processes contribute to the persistence and sustained replication of some bacterial viruses in aquatic environments.
| |
INTRODUCTION |
|---|
Numerous studies have demonstrated that significant numbers of viruses exist throughout the biosphere (2, 11, 24, 25, 34). Bacterial viruses have been hypothesized to play key roles in controlling bacterial host population density and diversity, nutrient flow in marine food webs, and the transfer of genes between hosts (9, 13, 25, 32, 33).
In aquatic environments several environmental factors, including host-cell starvation (26) and UV radiation exposure, substantially influence bacteriophage replication (13, 21, 22, 30, 31, 39). UV-C radiation (190 to 290 nm) is absorbed by nucleic acids, resulting in rapid viral inactivation (12). The UV-C component of solar UV radiation is removed by the stratospheric ozone layer, and no longer penetrates to the biosphere (16). In contrast, the longer-wavelength solar UV radiation bands (UV-A [320 to 400 nm] and UV-B [290 to 320 nm]) do penetrate the stratospheric ozone layer and substantially impact many ecosystems (7, 16). Solar UV exposure not only results in viral DNA damage and inactivation but also reduces the capacity of an infected host cell to support bacteriophage replication (16, 38).
The detrimental effects of solar UV exposure on bacteriophage persistence and replication make a capacity for viral DNA damage repair ecologically relevant. In order to partially explain the abundance of viruses in environments that receive substantial amounts of solar radiation, it has been hypothesized that viruses utilize several DNA repair mechanisms to allow for effective reproduction (10, 30, 37). Clearly, a capacity to either directly repair or possibly escape from solar UV radiation damage by executing an alternative replicative strategy such as lysogeny establishment would augment bacteriophage replication in aquatic environments with high radiation levels.
In this study, we report the isolation and partial characterization of a Pseudomonas aeruginosa bacteriophage, designated UNL-1, that possesses a novel capacity to reactivate DNA damage. This phenotype was expressed under conditions in which no previously described bacteriophage undergoes UV radiation-inducible or Weigle reactivation (36) and may contribute to virus replication and persistence under conditions of significant UV irradiation.
| |
MATERIALS AND METHODS |
|---|
Bacterial strains and bacteriophages. The bacterial strains
and bacteriophages used in this study are listed in Table 1. The R2 plasmid,
IncP-9 UVR (15), was obtained
from G. A. Jacoby. Bacteriophage UNL-1 was originally
isolated from a spontaneously appearing plaque (27).
TABLE 1. Bacterial strains and bacteriophages used in
this study
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
Media and chemicals. Cultures were grown in Luria-Bertani
broth (LB) (26).
Antibiotics were obtained from Sigma Chemical Corp. (St. Louis, Mo.).
Carbenicillin was used at a concentration of 300 µg ml![]() 1 to select for the presence
of the R2 plasmid in P. aeruginosa. Chloramphenicol was
used at a concentration of 250 µg/ml. TMN buffer (10 mM
Tris-HCl, pH 7.4; 10 mM MgSO4; 150 mM NaCl) was
used as phage diluent and as incubation medium for starved cells
used in one-step growth experiments. Cells for UV irradiation
experiments were suspended in saline (0.85% [wt/vol] NaCl).
1 to select for the presence
of the R2 plasmid in P. aeruginosa. Chloramphenicol was
used at a concentration of 250 µg/ml. TMN buffer (10 mM
Tris-HCl, pH 7.4; 10 mM MgSO4; 150 mM NaCl) was
used as phage diluent and as incubation medium for starved cells
used in one-step growth experiments. Cells for UV irradiation
experiments were suspended in saline (0.85% [wt/vol] NaCl).
One-step growth curve experiments. A modification of the method of Ellis and Delbrück (8) was used for these experiments. P. aeruginosa host cells were grown to exponential phase (60 Klett units at 660 nm [Klett660 units]) in LB at 37°C and infected with bacteriophage UNL-1 at a multiplicity of infection of 0.1. Infected cells were diluted 10,000-fold in LB growth medium, and titers of the growth tube were obtained periodically to quantify progeny bacteriophage. Experiments were repeated a total of three times. One-step growth experiments were performed with stationary-phase host cells by the method of Schrader et al. (26). In these experiments, host cells were starved by incubation in TMN buffer at 37°C for a period of 24 h prior to infection.
Estimation of the bacteriophage UNL-1 genome G/C ratio. The G/C composition of bacteriophage UNL-1 was determined by the method of Marmur and Doty (19) by using an AVIV Instruments (Lakewood, N.J.) model 14DS UV-Vis spectrophotometer equipped with a temperature coupler. Bacteriophage UNL-1 DNA was isolated by using a modification of the method of Miller et al. (20).
Generalized transduction experiments with bacteriophage UNL-1. The ability of bacteriophage UNL-1 to act as a generalized transducing virus was assessed by the method of Jensen et al. (17).
Bacteriophage inactivation with UV-C radiation.
Bacteriophage lysates were produced by using a modification of the soft agar
overlay method of Arber et al. (1). To quantify
bacteriophage inactivation by UV-C radiation, lysates were diluted
1,000-fold in TMN buffer, and 0.5-ml samples were placed in plastic
10-cm-diameter Petri dishes. Diluted bacteriophage samples were
exposed to UV-C radiation from a General Electric 15-W germicidal
lamp (G15T8) at a flux of 0.14 J m![]() 2 s
2 s![]() 1. The lamp radiation output
was determined with a UVX Radiometer (Ultra-Violet Products, San
Gabriel, Calif.) equipped with a UV-25 probe. The UV dose was varied
by exposing the samples to radiation for increasing time periods. The
percent inactivation of exposed bacteriophage lysates was determined
by plaque assay on unirradiated host cells as described by Simonson
et al. (28).
Bacteriophage lysates were irradiated with a UV-C dose sufficient to
inactivate 99% of the original PFU present in the unirradiated
lysate. All experimental manipulations involving UV-damaged viruses
were done under amber light to prevent photoreactivation.
1. The lamp radiation output
was determined with a UVX Radiometer (Ultra-Violet Products, San
Gabriel, Calif.) equipped with a UV-25 probe. The UV dose was varied
by exposing the samples to radiation for increasing time periods. The
percent inactivation of exposed bacteriophage lysates was determined
by plaque assay on unirradiated host cells as described by Simonson
et al. (28).
Bacteriophage lysates were irradiated with a UV-C dose sufficient to
inactivate 99% of the original PFU present in the unirradiated
lysate. All experimental manipulations involving UV-damaged viruses
were done under amber light to prevent photoreactivation.
Weigle reactivation procedures. (i) UV-C-irradiated host
cells. Weigle reactivation of UV-inactivated bacteriophages (36) was quantified
by using a modification of the procedure of Simonson et al. (28). The host cells
were grown to early exponential phase, harvested by centrifugation,
and suspended in an equal volume of saline. For UV-C exposure,
1 ml of suspended cells was added to each of 10-cm-diameter
petri dish, and the suspended cells were irradiated with germicidal
lamp UV-C radiation. The irradiated cells were added to the diluted
UV-C-irradiated bacteriophage samples, and the plaque formation
ability of the irradiated bacteriophages was quantified by plaque
assay. Due to the low survival of cells at the higher UV-C doses,
approximately 2 × 107 CFU of unirradiated cells were added
at the time of plating to the 10- and 20-J m![]() 2 samples. This allowed
formation of a confluent lawn and more-effective detection of phage
plaques. The Weigle reactivation factor was determined by using the
protocol of Simonson et al. (28). In
these experiments, the host cells were exposed to either UV-A
or UV-C radiation prior to infection and then mixed with
UV-C-irradiated bacteriophage under amber light illumination in which
photoreactivation of viral damage was not possible. This assay
quantifies the number of bacteriophages that were reactivated or
repaired by the host-cell dark repair mechanisms and consequently
enabled to produce plaques on host lawns after infection. The Weigle
reactivation factor is the ratio of phage titers obtained when host
cells are exposed to radiation in comparison to unirradiated
(control) host cells. Under conditions in which host cell irradiation
induces Weigle reactivation, the Weigle reactivation factor will be
increased as more viruses are repaired in the irradiated host cells
than in the unirradiated control hosts.
2 samples. This allowed
formation of a confluent lawn and more-effective detection of phage
plaques. The Weigle reactivation factor was determined by using the
protocol of Simonson et al. (28). In
these experiments, the host cells were exposed to either UV-A
or UV-C radiation prior to infection and then mixed with
UV-C-irradiated bacteriophage under amber light illumination in which
photoreactivation of viral damage was not possible. This assay
quantifies the number of bacteriophages that were reactivated or
repaired by the host-cell dark repair mechanisms and consequently
enabled to produce plaques on host lawns after infection. The Weigle
reactivation factor is the ratio of phage titers obtained when host
cells are exposed to radiation in comparison to unirradiated
(control) host cells. Under conditions in which host cell irradiation
induces Weigle reactivation, the Weigle reactivation factor will be
increased as more viruses are repaired in the irradiated host cells
than in the unirradiated control hosts.
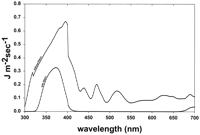
(ii) Solar UV-irradiated host cells. For experiments in which host cells were irradiated with UV-A radiation or sunlight, 2- to 5-ml samples of saline-suspended cells were added to glass petri dishes (32 mm in diameter), and the cells were maintained in suspension during irradiation by continuous agitation with a flea-sized stir bar. The petri dish lid was removed during the period of irradiation. UV-A radiation was generated with a 150-W Oriel Solar Simulator, model number 66002 (Oriel Corp., Stratford, Conn.), which passes solar UV radiation of approximately 280 to 400 nm plus residual longer wavelengths (Fig. 1). To obtain UV-A radiation in the absence of contaminating shorter-wavelength UV-B and UV-C range radiation, the Solar Simulator output was modified to eliminate UV-B and UV-C wavelengths by interposing two Oriel number 59459 B/C blocking filters that have a 320-nm cutoff and an Oriel number 59800 bandpass filter, which eliminates wavelengths of 400 to 700 nm and decreases the external transmittance of visible light by 50% (Fig. 1). With these filters in place, no UV radiation of less than 325 nm was transmitted to the samples. The UV-A fluence was quantified with a LI-COR (Lincoln, Nebr.) model Li-1800uw Underwater Spectroradiometer.
For experiments in which the host cells were exposed to sunlight, the sample in the petri dish was placed in direct sunlight with the glass lid removed, and samples were taken after various exposure times. The sunlight-irradiated cells were mixed with diluted UV-C-irradiated bacteriophage, and the amount of infective virus was quantified by plaque assay. All sunlight reactivation experiments were repeated three times and performed during the month of September.
Weigle reactivation experiments in which host cells were treated with chloramphenicol followed the same protocol detailed for the UV-A host cell irradiation experiments except that 250 µg of chloramphenicol mlPhotoreactivation of UV-C-irradiated bacteriophage UNL-1 lysates. The potential for photoreactivation of UV-irradiated UNL-1 lysates was quantified by using a modification of the method of Brash and Haseltine (4). Titers of irradiated lysates were obtained in quadruplicate on host cells. Two titration plates were incubated in a 37°C incubator at a distance of 45 cm from two 15-W GE Cool White bulbs (photoreactivating conditions), while the other set of plates was incubated under dark conditions in which photoreactivation was not possible.
Weigle reactivation experiments that were conducted under conditions in which photoreactivation was possible were performed by following the protocol described for solar UV-irradiated hosts except the plates were incubated in a 37°C incubator at a distance of 45 cm from two 15-W GE Cool White bulbs.Southern blot analysis of bacteriophage DNA. Bacteriophage DNA was isolated by using a modification of the method of Miller et al. (20). Restriction endonucleases were obtained from New England Biolabs (Beverly, Mass.) and used under the conditions recommended by the manufacturer. Bacteriophage DNA samples were digested with EcoRV, and the resulting fragments were separated on a 0.8% (wt/vol) agarose gel and transferred to a Nytran membrane (Schleicher & Schuell, Keene, N.H.) by using the Turbo Blotter rapid downward transfer system (Midwest Scientific, Valley Park, Mo.). Bacteriophage DNA probe was generated by using a Random Prime labeling kit (Amersham Life Sciences, Arlington Heights, Ill.) and visualized by using chemiluminescence direct imaging as specified by the manufacturer.
| |
RESULTS |
|---|
Bacteriophage UNL-1 virion morphology and classification. Electron micrographic examination revealed that the UNL-1 virion was structurally complex (Fig. 2). The morphology of UNL-1 most closely resembled the Myoviridae family (6) of large and structurally complex bacterial viruses that possess contractile tails with fibers. It had an icosahedral capsid that was approximately 80 nm in diameter, with a sheathed, contractile tail with a total length of approximately 200 nm. The sheath began 10 nm from the capsid, was approximately 80 nm in length, and terminated in a baseplate with attached tail fibers.

UNL-1 nucleic acid analyses. The G/C content of the UNL-1 genome was determined to be approximately 68%. UNL-1 DNA was restricted by EcoRV but was not restricted by endonucleases BamHI, SalI, or XhoI. Based on the EcoRV restriction fragment banding pattern, the size of the UNL-1 chromosome was estimated to be approximately 48 kb. The EcoRV restriction pattern of UNL-1 was compared to the P. aeruginosa Siphoviridae family bacteriophages D3, F116, and F116d1; a variant of bacteriophage F116, G101; and the Myoviridae family virus UT1; as well as the Escherichia coli bacteriophages T4 (Myoviridae) and phage lambda (Siphoviridae). P. aeruginosa bacteriophage UT1 DNA when digested with EcoRV produced a restriction pattern similar to that of EcoRV-digested UNL-1 (data not shown). Southern blot analysis with UNL-1 DNA as a probe revealed a substantial degree of base sequence identity with P. aeruginosa Myoviridae family bacteriophage UT1 DNA under conditions of high stringency (Fig. 3). Under the conditions employed in this experiment, a minimum of 86% DNA base sequence identity is required to produce a hybridization signal. No UNL-1 probe hybridization was detected to E. coli bacteriophages T4 or lambda or to the other P. aeruginosa bacteriophage DNA fragments under these experimental conditions.

Bacteriophage UNL-1 one-step growth curve. A one-step growth curve (8) experiment was performed by using a mid-logarithmic-phase P. aeruginosa PAO303 host culture grown in LB at 37°C with aeration. UNL-1 had a mean burst size of 380 PFU and a latent period of approximately 65 min under these conditions. UNL-1 replication in stationary-phase hosts was assessed by performing one-step growth experiments in P. aeruginosa PAO303 hosts by the procedures of Schrader et al. (26). UNL-1 replication was not detected in P. aeruginosa PAO303 cells that were starved for 24 h at 37°C prior to infection under the conditions and time periods employed in our experiments (data not shown).
Bacteriophage UNL-1 does not mediate generalized transduction. Bacteriophage UT1 is a lytic, generalized transducing virus of P. aeruginosa originally isolated from Fort Loudoun Lake, Tenn. (23). Because of the substantial DNA base sequence identity revealed by the Southern blot experiments between UNL-1 and UT1, the ability of bacteriophage UNL-1 to act as a generalized transducing virus was examined by the method of Jensen et al. (17). Using a range of bacteriophage multiplicities of infection, reversions to arginine prototrophy in recipient P. aeruginosa PAO303 cell populations were not detected at rates greater than those of uninfected controls (data not shown). Despite the DNA sequence relationship to UT1 revealed in restriction fragment pattern comparisons and Southern blot experiments, these experiments revealed that UNL-1 was either a nontransducing bacteriophage or that transduction with this virus was substantially less efficient than that exhibited by bacteriophage UT1 under the same experimental conditions.
UV-A irradiation of host cells results in the induction of bacteriophage UNL-1 reactivation. Repair of viral DNA damage was quantified by Weigle reactivation experiments. PAO303 host cells were exposed to UV-A radiation and infected with UV-C-irradiated UNL-1 or G101 bacteriophages. An approximately 13-fold induction of UNL-1 plaque-forming ability was observed in UV-A-irradiated host cells (Fig. 4A). In contrast, under the identical experimental conditions, Weigle reactivation induction was not evident for UV-C-damaged bacteriophage G101 (Fig. 4A).

In order to determine if the UV-A-inducible UNL-1 reactivation was dependent on the presence of a functional RecA protein, the recA deletion strain, P. aeruginosa PDO2, was irradiated with UV-A radiation and used as the host for Weigle reactivation experiments (Fig. 4B). Weigle reactivation induction of up to 13-fold was also observed with UV-C-damaged UNL-1 in the recA host, while no detectable reactivation induction was observed with UV-C-damaged G101 when the host lacked a functional RecA protein (Fig. 4B).
Exposure of host cells to UV-C radiation fails to induce UNL-1 Weigle reactivation. UV-C irradiation of host cells failed to induce Weigle reactivation of UV-C-damaged bacteriophage UNL-1 (Fig. 5). In addition, no Weigle reactivation induction of UV-damaged bacteriophage G101 was evident under identical experimental conditions. UV-C-irradiated host cells containing the UV resistance plasmid R2 supported only an approximately fourfold reactivation induction for UV-damaged G101 (Fig. 5). After UV-C irradiation, identical R2 plasmid-containing hosts did not support enhanced UNL-1 reactivation (Fig. 5).

Exposure of host cells to sunlight induces UNL-1 reactivation. P. aeruginosa PAO303 host cells were exposed to sunlight and used in Weigle reactivation experiments. Sunlight-irradiated host cells were removed to the laboratory and infected with UV-damaged bacteriophages only under amber light illumination where photoreactivation of the infecting damaged bacteriophage was not possible. Exposing host cells to sunlight resulted in an eightfold reactivation induction of UV-damaged bacteriophage UNL-1 (Fig. 6).

Host cell pretreatment with chloramphenicol blocks UV-A induced UNL-1 reactivation. To investigate the requirement for host cell protein synthesis for UV-A induction of reactivation, cells were treated with chloramphenicol prior to UV-A exposure as described in Materials and Methods, harvested by centrifugation, and suspended in saline for irradiation. Treatment of the cells with chloramphenicol resulted in the suppression of UV-A-induced UNL-1 Weigle reactivation induction (Fig. 7).

UV-C irradiation of bacteriophage UNL-1 virions produces damage that is photoreactivatable. To confirm that a portion of the lethal damage acquired by UNL-1 due to UV-C irradiation was due to the production of cyclobutane pyrimidine dimers, UV-C-irradiated UNL-1 was photoreactivated by using a modification of the method of Brash and Haseltine (4). UV-C-damaged bacteriophage UNL-1 showed an average of 15-fold-greater plaque formation ability in PAO303 incubated under conditions in which photoreactivation was possible in comparison to the same host cells incubated in the dark (Table 2).
Photoreactivation does not abolish UV-A inducible Weigle reactivation of UNL-1. To assess whether photoreactivation decreased UNL-1 Weigle reactivation, experiments were conducted under conditions in which photoreactivation was possible. Under these conditions, an approximately 9.6-fold induction of UNL-1 reactivation was still observed in UV-A-irradiated cells with the same kinetics as the dark-incubated cells (Fig. 8), suggesting that photoreactivation does not completely eliminate the DNA damage that is repaired by the UV-A inducible repair system of UNL-1. Bacteriophage G101 reactivation increased approximately fourfold under these conditions.

| |
DISCUSSION |
|---|
Our experiments revealed that P. aeruginosa bacteriophage UNL-1 was a lytic, nontransducing bacteriophage. Electron micrographic examination revealed that the virion had a complex structure and most closely resembled bacteriophages in the Myoviridae family. Under conditions of rapid host growth, bacteriophage UNL-1 displayed a latent period of approximately 65 min, with a mean burst size of 380. Agarose gel electrophoresis of restricted UNL-1 DNA demonstrated that the chromosome was approximately 48 kb in size, and Southern blotting experiments revealed a substantial level of DNA sequence identity with the previously described P. aeruginosa bacteriophage UT1. While similar to UT1 at the DNA sequence level, generalized transduction was not detected in experiments with bacteriophage UNL-1.
Bacteriophage UNL-1 exhibited a unique capacity for UV damage reactivation. Weigle reactivation is a phenomenon whereby inducible host cell DNA repair systems respond to UV irradiation and eliminate damage from the chromosomes of both the irradiated host and infecting UV-C-inactivated bacteriophages, ultimately allowing replication of the now-repaired virus and the death of the host cell (12, 36). After infecting a UV-A-irradiated host cell, irradiated bacteriophage UNL-1 was able to undergo inducible dark repair and to recover infectivity. Exposure of the host cells to chloramphenicol abolished this reactivation induction (Fig. 7) and revealed that host cell protein synthesis was required after UV-A exposure for the expression of UNL-1 reactivation induction.
The reactivation of UV-damaged UNL-1 was clearly distinct from all previously described instances of bacteriophage Weigle reactivation based on the following evidence. (i) UNL-1 reactivation was induced by broad-band UV-A exposure to the host cell (Fig. 4) and was not enhanced by host cell UV-C irradiation (Fig. 5). In contrast, all previously described virus Weigle reactivation systems are activated by UV-C irradiation of the host and are unresponsive to longer-wavelength UV-A radiation exposure (7). (ii) Bacteriophage UNL-1 UV-A-inducible reactivation was not dependent on the host recA gene product. Substantial induction of reactivation was evident in the P. aeruginosa recA deletion mutant strain PDO2 (Fig. 5). All other previously described Weigle UV damage reactivation and SOS repair systems are completely dependent on a functional host recA gene to initiate induction (35). (iii) Bacteriophage UNL-1 Weigle reactivation was inducible in P. aeruginosa PAO303, a host that has been previously demonstrated to lack any UV-inducible Weigle reactivation capabilities unless the host bacterium also contained the UV resistance plasmid R2 (28). Additional experiments with bacteriophage D3 have confirmed that this virus, like G101, is incapable of undergoing UV-A inducible Weigle reactivation in P. aeruginosa PAO303 hosts (27). (iv) UNL-1 is incapable of reactivation induction in the presence of the plasmid R2. While experiments to assess whether the UNL-1 virus could supply reactivation functions in trans to superinfecting UV-damaged bacteriophage were not performed due to the lack of suitable UNL-1 mutants and the potential for UNL-1 interference with the replication of superinfecting, heterologous bacteriophages, the simplest hypothesis to account for all of these observations is that bacteriophage UNL-1 encodes a gene(s) not present on other bacteriophages, allowing for expression of Weigle reactivation in UV-A-irradiated hosts.
To further characterize the repair mechanism, photoreactivation studies were undertaken to elucidate the DNA damage that is repaired by UNL-1. DNA exposure to UV-C radiation results in the formation of cyclobutane pyrimidine dimers with a high efficiency (12). Evidence for the existence of cyclobutane pyrimidine dimers in cell or virus DNA is the ability to reverse the lethal effects of DNA damage by exposure to photoreactivating light (12). When P. aeruginosa host cells were infected with UV-irradiated bacteriophage UNL-1 and incubated under conditions in which photoreactivation was possible, a nearly 15-fold increase in plaque-forming ability was observed in comparison to samples of the same infected cells incubated in the dark. These experiments revealed that cyclobutane pyrimidine dimers were created by UV-C irradiation of bacteriophage UNL-1 and that this class of DNA damage was subject to reversal by host cell photoreactivation. Whether cyclobutane pyrimidine dimers are actually repaired or bypassed in UV-A-irradiated host cells infected with UV-damaged bacteriophage UNL-1 when incubated in the dark is not firmly established and is under investigation. The observation that under conditions in which photoreactivation was possible a substantial UV-A induction of UNL-1 reactivation was still evident (Fig. 8) suggests that the lesion repaired by the UV-A-inducible system is not exclusively photoreactivatable pyrimidine dimers. If photoreactivation and the UV-A-inducible dark-repair systems reversed precisely the same lesions, the incubation of damaged bacteriophages under conditions allowing for photoreactivation would be expected to sharply decrease the detectable levels of Weigle reactivation. However, it is possible that the UV-A-inducible dark-repair system acting on UNL-1 actually does reverse precisely the same lesions as the photoreactivation, but it does so either slowly or incompletely. Further biochemical characterization of the function(s) involved in UNL-1 reactivation will be required to resolve this point.
Sunlight exposure is a major factor in the destruction of viral infectivity
in aquatic ecosystems (22, 31, 39). Exposure
of host cells to sunlight also induced Weigle reactivation of
UV-irradiated UNL-1 (Fig. 6). UV-B
wavelengths, the most damaging of the solar wavelengths, have an
attenuation coefficient of 0.86 m![]() 1 in moderately productive
coastal marine waters (21, 29). At
a depth of 10 m, approximately 2% of the surface UV-B radiation
is still present. The longer wavelengths of UV-A radiation are
able to penetrate to significantly greater depths. These facts
suggest that in ecosystems exposed to UV radiation, bacteriophages
able to express a UV-A-inducible reactivation activity would have
opportunities to reactivate DNA damage. Moreover, the differential
penetration ability of UV wavelengths suggests that in deep systems,
UV-A-inducible systems could be highly active and that the
elimination of DNA damage would be efficient in a large portion of
aquatic ecosystems. Weinbauer et al. (37) suggest active
photoreactivation as the most reasonable explanation for the
conspicuously high virus titers consistently observed in aquatic
systems exposed to high UV radiation levels. The observation of the
UV-A-inducible DNA damage repair potential of UNL-1 suggests that
additional mechanisms may contribute to the maintenance of viral
infectivity in environments with substantial levels of UV radiation
as well.
1 in moderately productive
coastal marine waters (21, 29). At
a depth of 10 m, approximately 2% of the surface UV-B radiation
is still present. The longer wavelengths of UV-A radiation are
able to penetrate to significantly greater depths. These facts
suggest that in ecosystems exposed to UV radiation, bacteriophages
able to express a UV-A-inducible reactivation activity would have
opportunities to reactivate DNA damage. Moreover, the differential
penetration ability of UV wavelengths suggests that in deep systems,
UV-A-inducible systems could be highly active and that the
elimination of DNA damage would be efficient in a large portion of
aquatic ecosystems. Weinbauer et al. (37) suggest active
photoreactivation as the most reasonable explanation for the
conspicuously high virus titers consistently observed in aquatic
systems exposed to high UV radiation levels. The observation of the
UV-A-inducible DNA damage repair potential of UNL-1 suggests that
additional mechanisms may contribute to the maintenance of viral
infectivity in environments with substantial levels of UV radiation
as well.
The evidence that virus-encoded, host-independent DNA repair systems exist in eukaryotic algal viruses (10), as well as in prokaryotic bacterial viruses (40), supports the hypothesis that DNA repair systems are evolutionarily ancient and, as a consequence, have a significant and widespread distribution in the biosphere (3), including the viruses. The discovery of a novel reactivation capacity expressed by bacteriophage UNL-1 suggests that both host and virus DNA repair systems act in concert to play an important role in virus replication and the maintenance of viral infectivity in aquatic ecosystems.
| |
ACKNOWLEDGMENTS |
|---|
This research was supported in part by Cooperative Agreement CR822163 with the U.S. Environmental Protection Agency, Cooperative Agreement 9255225 from the National Science Foundation EPSCoR program, the University of Nebraska-Lincoln Water Center, and Midwestern University.
We thank M. Griep for help with the UNL-1 G/C analysis and K. W. Nickerson for critical evaluation of the manuscript.
| |
FOOTNOTES |
|---|
* Corresponding author. Mailing address: Arizona College of Osteopathic Medicine, Midwestern University, 19555 N. 59th Ave., Glendale, AZ 85308. Phone: (602) 572-3225. Fax: (602) 572-3226. E-mail: tkokjo@arizona.midwestern.edu.
| |
REFERENCES |
|---|
| 1. | Arber, W., L. Enquist, B. Hohn, N. E. Murray, and K. Murray. 1983. Experimental methods for use with lambda, p. 433-466. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. |
| 2. | Bergh, O., K. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468[Medline]. |
| 3. | Bernstein, H., and C. Bernstein. 1989. Bacteriophage T4 genetic homologies with bacteria and eukaryotes. J. Bacteriol. 171:2265-2270[Medline]. |
| 4. | Brash, D. E., and W. A. Haseltine. 1985. Photoreactivation of Escherichia coli reverses umuC induction by UV light. J. Bacteriol. 163:460-463[Medline]. |
| 5. | Cavenagh, M. M., and R. V. Miller. 1986. Specialized transduction Pseudomonas aeruginosa PAO by bacteriophage D3. J. Bacteriol. 165:448-452[Medline]. |
| 6. | Coetzee, J. N. 1987. Bacteriophage taxonomy, p. 45-85. In S. M. Goyal, C. P. Gerba, and G. Bitton (ed.), Phage ecology. John Wiley & Sons, Inc., New York, N.Y. |
| 7. | Eisenstark, A. 1989. Bacterial genes involved in response to near-ultraviolet radiation. Adv. Genet. 26:99-147[Medline]. |
| 8. | Ellis, E. L., and M. Delbrück. 1939. The growth of bacteriophage. J. Gen. Physiol. 22:365-384. |
| 9. | Fuhrman, J. A., and C. A. Suttle. 1993. Viruses in marine planktonic systems. Oceanography 6:51-63. |
| 10. | Furuta, M., J. O. Schrader, H. S. Schrader, T. A. Kokjohn, S. Nyaga, A. K. McCullough, R. S. Lloyd, D. E. Burbank, D. Landstein, L. Lane, and J. L. Van Etten. 1997. Chlorella virus PBCV-1 encodes a homolog of the bacteriophage T4 UV damage repair gene denV. Appl. Environ. Microbiol. 63:1551-1556[Abstract]. |
| 11. | Hara, S., I. Koike, K. Terauchi, H. Kamiya, and E. Tanoue. 1996. Abundance of viruses in deep oceanic waters. Mar. Ecol. Prog. Ser. 145:269-277. |
| 12. | Harm, W. 1980. Biological effects of ultraviolet radiation. Cambridge University Press, New York, N.Y. |
| 13. | Heldal, M., and G. Bratbak. 1991. Production and decay of viruses in aquatic environments. Mar. Ecol. Prog. Ser. 72:205-212. |
| 14. | Horn, J. M., and D. E. Ohman. 1988. Transcriptional and translational analyses of recA mutant alleles in Pseudomonas aeruginosa. J. Bacteriol. 170:1637-1650[Medline]. |
| 15. | Jacoby, G. A. 1977. Classification of
plasmids in Pseudomonas aeruginosa, p. 119-126. In D.
Schlessinger (ed.), Microbiology |
| 16. | Jagger, J. 1985. Solar-UV actions on living cells. Praeger Publishers, New York, N.Y. |
| 17. | Jensen, E. C., H. S. Schrader, B. Rieland, T. L.
Thompson, K. W. Lee, K. W. Nickerson, and T. A. Kokjohn. 1998.
Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus
natans, Escherichia coli, and Pseudomonas aeruginosa.
Appl. Environ. Microbiol. 64:575-580 |
| 18. | Krishnapillai, V. 1971. A novel transducing phage. Its role in recognition of a possible new host controlled modification system in Pseudomonas aeruginosa. Mol. Gen. Genet. 114:134-143. |
| 19. | Marmur, J., and P. Doty. 1962. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 5:109-118. |
| 20. | Miller, R. V., J. M. Pemberton, and K. E. Richards. 1974. F116, D3, and G101: temperate bacteriophages of Pseudomonas aeruginosa. Virology 59:566-569[Medline]. |
| 21. | Murray, A. G., and G. A. Jackson. 1993. Viral dynamics II. A model of the interaction of ultraviolet light and mixing processes on virus survival in seawater. Mar. Ecol. Prog. Ser. 102:105-114. |
| 22. | Noble, R. T., and J. A. Fuhrman. 1997. Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 63:77-83[Abstract]. |
| 23. | Ogunseitan, O. A., G. S. Sayler, and R. V. Miller. 1990. Dynamic interactions between Pseudomonas aeruginosa and bacteriophages in lakewater. Microb. Ecol. 19:171-185. |
| 24. | Paul, J. H., S. C. Jiang, and J. M. B. Rose. 1991. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl. Environ. Microbiol. 57:2197-2204[Medline]. |
| 25. | Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. |
| 26. | Schrader, H. S., J. O. Schrader, J. J. Walker, T. A. Wolf, K. W. Nickerson, and T. A. Kokjohn. 1997. Bacteriophage infection and multiplication occur in Pseudomonas aeruginosa starved for 5 years. Can. J. Microbiol. 43:1157-1163[Medline]. |
| 27. | Schrader, J. O. 1997. The role of extra-chromosomal elements in ultraviolet resistance in Pseudomonas aeruginosa. M.S. thesis. University of Nebraska-Lincoln, Lincoln. |
| 28. | Simonson, C. S., T. A. Kokjohn, and R. V. Miller. 1990. Inducible UV repair potential of Pseudomonas aeruginosa PAO. J. Gen. Microbiol. 136:1241-1249[Medline]. |
| 29. | Smith, R. C., and K. S. Baker. 1979. Penetration of UV-B and biologically effective dose-rates in natural waters. Photochem. Photobiol. 29:311-323. |
| 30. | Suttle, C. A., A. M. Chan, F. Chen, and D. R. Garza. 1993. Cyanophages and sunlight: a paradox, p. 303-308. In R. Guerrero, and C. Pedros-Alio (ed.), Trends in microbial ecology. Spanish Society for Microbiology, Barcelona, Spain. |
| 31. | Suttle, C. A., and F. Chen. 1992. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58:3721-3729[Abstract]. |
| 32. | Suttle, C. A. 1994. The significance of viruses to mortality in aquatic microbial communities. Microbial. Ecol. 28:237-243. |
| 33. | Thingstad, T. F., M. Heldal, G. Bratbak, and I. Dundas. 1993. Are viruses important partners in pelagic food webs? Trends Evol. Ecol. 8:209-213. |
| 34. | Torrella, F., and R. Y. Morita. 1979. Evidence by electron micrographs for a high incidence of bacteriophage particles in the waters of Yaquina Bay, Oregon: ecological and taxonomical interpretations. Appl. Environ. Microbiol. 37:774-778[Medline]. |
| 35. | Walker, G. C. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 48:60-93. |
| 36. | Weigle, J. J. 1953. Induction of mutation in a bacterial virus. Proc. Natl. Acad. Sci. USA 39:628-636. |
| 37. | Weinbauer, M. G., S. W. Wilhelm, C. A. Suttle, and D. R. Garza. 1997. Photoreactivation compensates for UV damage and restores infectivity to natural marine virus communities. Appl. Environ. Microbiol. 63:2200-2205[Abstract]. |
| 38. | Wingo, R. J., T. Fossum, S. Luo, and J. Jagger. 1980. Reduction by near-ultraviolet (334 nm) light of Escherichia coli capacity for phage growth depends upon the rel gene and 4-thiouridine. Photochem. Photobiol. 32:353-360[Medline]. |
| 39. | Wommack, K. E., R. T. Hill, T. A. Muller, and R. R. Colwell. 1996. Effects of sunlight on bacteriophage viability and structure. Appl. Environ. Microbiol. 62:1336-1341[Abstract]. |
| 40. | Yasuda, S., and M. Sekiguchi. 1970. T4 endonuclease involved in repair of DNA. Proc. Natl. Acad. Sci. USA 67:1839-1845[Medline]. |
This article has been cited by other articles:
|
| |||||||||
| J. Bacteriol. | Microbiol. Mol. Bio. Review | Eukaryot. Cell | All ASM Journals |
|---|